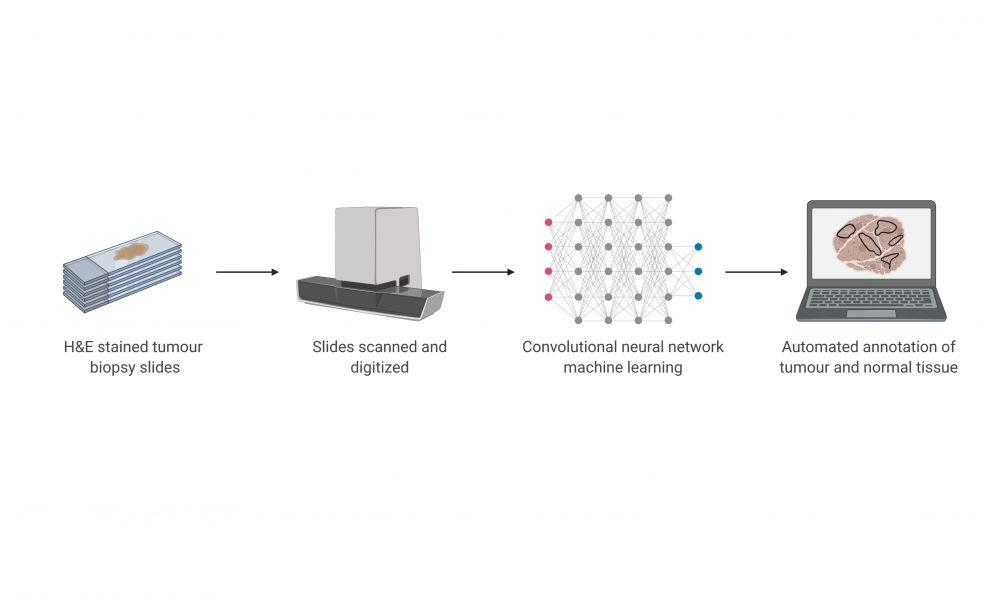
Significant advancements in sequencing technologies have enabled rapid genomic and molecular characterization of cancers, improving diagnostics and treatment planning. The annotation of tumour tissue samples prior to these analyses, however, still requires the time and expertise of a pathologist and represents a bottleneck in cancer genomics.
In a new study published in the Journal of Pathology Informatics, led by GSC Distinguished Scientist Dr. Aly Karsan, researchers described a deep learning-based approach to distinguish tumour from normal tissue using digitized slides. The high accuracy of their automated method compared to manually conducted pathologist annotation suggests that automated image processing may enhance and expedite downstream analyses.
Tissue-based diagnostics are often conducted by staining and examining tissues under a microscope, enabling pathologists to determine the cancer stage and the size and number of tumours in a specimen. For downstream genomic analyses, pathologists will also distinguish the regions of the sample containing cancer from those which contain normal tissue. Tumour tissue can then be separated from the rest of the sample, enabling comparisons between the cancer and normal genomes.
Manual annotation of tissue specimens is labour intensive and suffers from poor reproducibility, with significant variation in results between pathologists. This led Dr. Karsan and researchers from GenerationsE Software Solutions, the Department of Pathology at the University of British Columbia, and Kaohsiung Medical University in Taiwan to investigate the utility of deep learning-based methods for colorectal cancer detection and segmentation from digitized histology slides.
Deep learning has been successfully applied to medical image processing, such as magnetic resonance imaging, computed tomography, biopsy and endoscopy. Recently, a convolutional neural network architecture was trained for the automated detection and classification of breast cancer metastases in whole-slide images of lymph node sections.
The research team took a similar approach to identify tumour regions in a model of colorectal cancer and compared the results to annotations performed by a pathologist. The approach achieved an accuracy of 99.9 per cent for normal slides and 94.8 per cent for cancer slides, suggesting that automation through the use of neural network algorithms may reduce the burden on pathologists and enhance confidence in colorectal cancer diagnostics and genomic analyses.
Lin Xu, Blair Walker, Peir-In Liang, Yi Tong, Cheng Xu, Yu Chun Su, Aly Karsan. 2020. Colorectal cancer detection based on deep learning. J Pathol Inform. 11:28.
The study was financially supported by the NRC-IRAP research grants.
Learn more about research in Dr. Karsan’s lab
Learn more about research at the GSC